Current research
Parkinson's disease affects 2% of the population aged over 65,
making it not only the most common movement disorder
but also the second most common neurodegenerative disease
(after Alzheimer's disease).
Parkinson's disease is believed to be caused by
the formation of protein aggregates in the brain,
by a protein called α-synuclein.
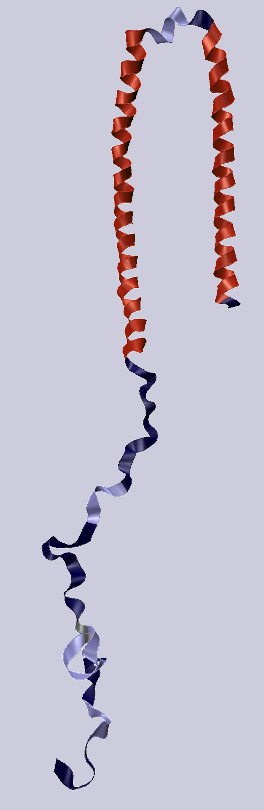
α-synuclein is an intrinsically disordered protein
that has the ability to change its secondary structure
depending on the environment.
Thus, its segments are disordered when surrounded by a solvent,
curl into α-helices near a membrane
or stretch into β-sheets in fibirls or amyloids.
This behavior is shared by a class of amyloid-forming proteins,
suggesting generic behavior.
We are investigating
the formation and structure of α-synuclein aggregates
by means of computer simulations.
To achieve this,
we are developing a highly coarse grained model
by representing segments of 10-15 aminoacids
as soft rigid bodies of various shapes
with attractive patches on their surfaces.
To simulate dynamics we have implemented a Brownian Dynamics algorithm
for the translational and rotational motion.




 Parkinson's disease affects 2% of the population aged over 65,
making it not only the most common movement disorder
but also the second most common neurodegenerative disease
(after Alzheimer's disease).
Parkinson's disease is believed to be caused by
the formation of protein aggregates in the brain,
by a protein called α-synuclein.
α-synuclein is an intrinsically disordered protein
that has the ability to change its secondary structure
depending on the environment.
Thus, its segments are disordered when surrounded by a solvent,
curl into α-helices near a membrane
or stretch into β-sheets in fibirls or amyloids.
This behavior is shared by a class of amyloid-forming proteins,
suggesting generic behavior.
Parkinson's disease affects 2% of the population aged over 65,
making it not only the most common movement disorder
but also the second most common neurodegenerative disease
(after Alzheimer's disease).
Parkinson's disease is believed to be caused by
the formation of protein aggregates in the brain,
by a protein called α-synuclein.
α-synuclein is an intrinsically disordered protein
that has the ability to change its secondary structure
depending on the environment.
Thus, its segments are disordered when surrounded by a solvent,
curl into α-helices near a membrane
or stretch into β-sheets in fibirls or amyloids.
This behavior is shared by a class of amyloid-forming proteins,
suggesting generic behavior.

